- Indico style
- Indico style - inline minutes
- Indico style - numbered
- Indico style - numbered + minutes
- Indico Weeks View
Advances and Challenges in Zebrafish Image and Video Analysis
Data controller
Anna-Christina Jauch Deutsches Elektronen-Synchrotron DESY
datenschutz@desy.de
Privacy notice
→
Europe/Berlin
T-Werk Probebühne (Schinkelhalle Potsdam)
T-Werk Probebühne
Schinkelhalle Potsdam
Schiffbauergasse 4i
14467 Potsdam
Description
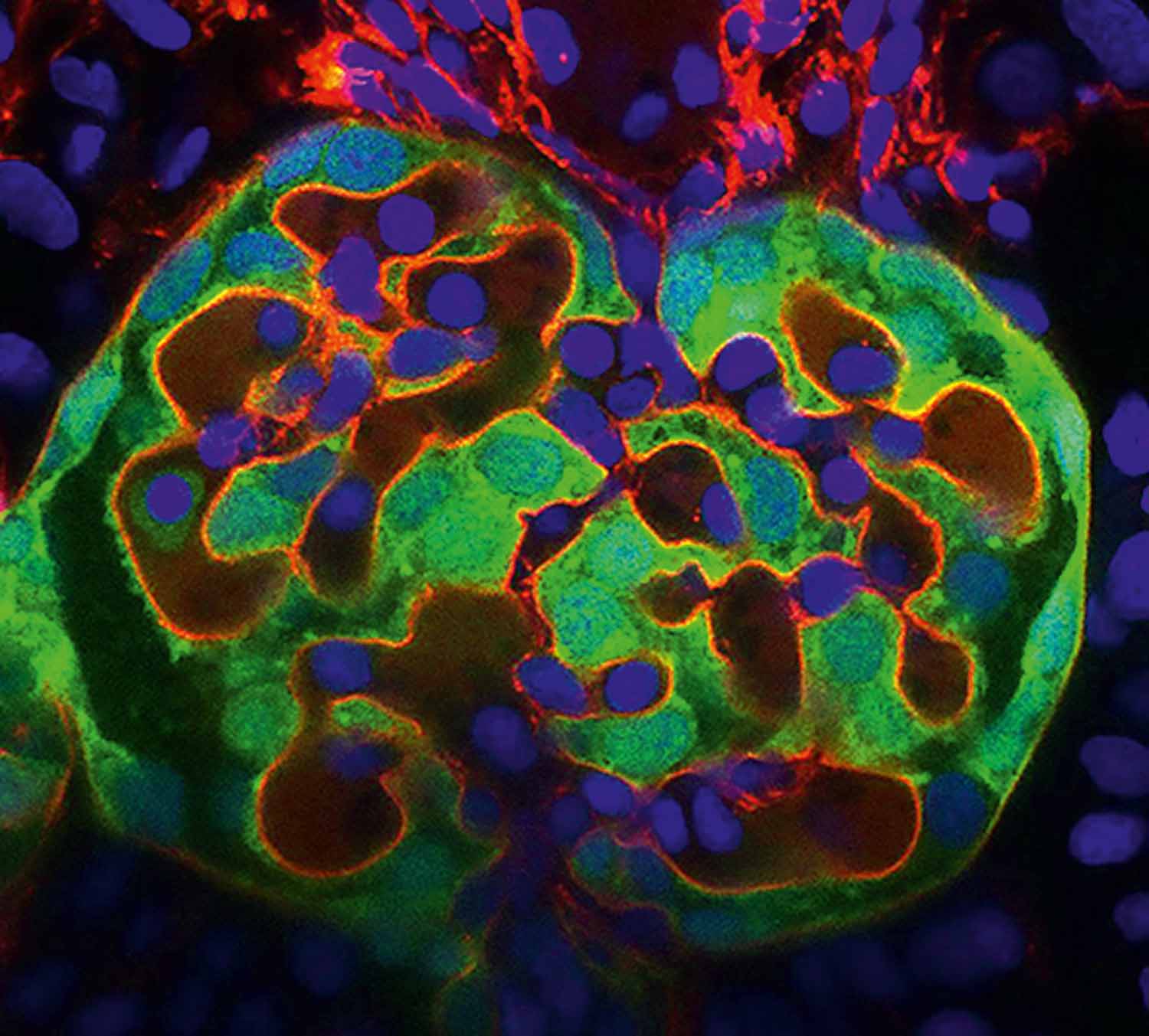
Image: Aikaterina Kourpa, MDC
This workshop aims to bring together researchers working with zebrafish to discuss recent advances and challenges in image and video analysis. Participants will have the opportunity to present their work in short oral presentations, share challenges they face, and discuss methods, tools, and approaches that could help improve zebrafish imaging workflows. Topics will also include strategies for storing, archiving, and organizing image and video data in line with FAIR principles.
Following the presentations, a roundtable discussion will provide an opportunity to explore potential joint activities, required tools, and supporting infrastructures to facilitate zebrafish imaging analyses.
R E G I S T R A T I O N I S O P E N N O W ! |
This workshop is planned as a pre-event for the Helmholtz Imaging Conference 2025. Participation in the Zebrafish workshop alone is possible and free of charge; however, we highly recommend experiencing the full conference. We invite you to register for all three days. You can find the conference program here.
Call for Abstracts
We invite you to actively contribute by submitting an abstract for a short oral presentation. Presentations should focus on:
- Current work involving zebrafish image or video analysis
- Challenges faced in analysis and data management
- Methods for storing, archiving, and processing image and video data in accordance with FAIR principles
- Future needs and tools that could facilitate zebrafish research
Submit your contribution via the Call for Abstracts form on the conference page. Do not forget to choose “Zebrafish” as one of the Keywords. Only in this way we can assign your contribution to the Workshop.
Please note that travel and accommodation costs are at your own expense.
The Zebrafish Workshop is jointly organized by the Helmholtz Centre for Environmental Research – UFZ/Dpt. Ecotoxicology, Biomedical Engineering & Robotics (BER) at KIT and Helmholtz Imaging.
BACK to the Conference Website
-
-
09:00
→
09:05
Welcome & IntroductionConveners: Christian Pylatiuk, Stefan Scholz (UFZ - ETOX)
-
09:05
→
10:10
Talks 1-5
-
09:05
Integration of live-imaging with single-cell genomics to dissect spatiotemporal gene regulation and its impact on cell behavior in the developing zebrafish embryo 13m
Morphogenesis during embryonic development relies on the continuous integration of complex spatiotemporal cues into gene regulatory consequences. Our lab uses zebrafish embryos as an optically accessible and genetically manipulatable model to investigate the link between gene regulation and dynamic cellular behaviors in vivo. We combine live-imaging with advanced genomics—including spatial transcriptomics, single-cell RNA sequencing, and epigenetic sequencing techniques—to dissect how cell-intrinsic programs are shaped by extrinsic cues.
The core expertise of our lab lies in the development of single-cell and spatial genomic sequencing methods, a technical repertoire that is inherently disruptive to the biological sample. Traditionally, transcriptomic readouts provide static snapshots of RNA expression, lacking both spatial and temporal resolution. A central aim of our research is to overcome these limitations by capturing space and time in genomic data. Lineage-tracing approaches allow us to reconstruct cell specification trajectories and quantify clonal diversity. Slam-seq time-stamps newly transcribed RNA molecules, encoding temporal information into transcriptomes. In parallel, we are advancing spatial RNA sequencing techniques to achieve increasingly refined spatial resolution of gene expression. While it remains technically impossible to continuously observe transcription in vivo, our long-term goal is to bridge this gap by integrating live-microscopy with genomic profiling.
One of these newly-initiated projects will focus on neural crest cells, a highly migratory, multipotent cell population that travels through diverse environments before integration to various, distinct tissues. By combining live-imaging approaches with time-resolved spatial transcriptomics, we will map their migratory paths alongside their changing transcriptional landscapes. This allows us to understand how transient interactions of the cell with surrounding microenvironment drive gene regulatory changes that will influence fate decisions and differentiation.
A second research direction investigates how cells sense and integrate mechanical inputs to drive morphogenic outcomes. We are interested in how distinct or converging mechanosensitive pathways are activated by external stimuli, translated into gene expression programs that influence cell morphology, migration and proliferation. To dissect these processes at a single-cell level, we use transplantation-based chimeric embryos and will in future also incorporate optogenetic tools to perturb signaling pathways with high spatial and temporal control. This approach will enable us to distinguish cell-autonomous effects from those driven by the tissue environment.
A major challenge across both projects is the analysis and integration of the complex imaging data with our genomic analyses. The segmentation and tracking of populations and/or single cell across time points in the growing environment of the developing embryo, is our current bottleneck. Therefore, we are actively seeking collaborations in computational image analysis and further modeling after integration of the multimodal datasets.Speaker: Anna Klaus -
09:18
Highly-multiplexed smFISH of sectioned zebrafish embryos 13m
The zebrafish embryo is a well-established and powerful in vivo system for both imaging and high-throughput genomics, yet applying cutting-edge approaches for spatial transcriptomics remains a challenge due to protocols, panels, and pipelines being generally focused on larger mammalian tissues. To meet this challenge, we developed an approach to allow higher-throuput microtome sectioning of multiple, spatially aligned, zebrafish embryos. The approach can be adapted to different developmental stages, spatial orientations, and throughput scenarios. We have performed highly multiplexed single-molecule RNA FISH using the 10x Genomics Xenium platform with a 300-gene custom probe panel on >30 serial sections of multiple spatially-aligned 24hpf zebrafish embryo tails. The custom gene panel was chosen mostly based on cell-type marker genes from our own 10x Genomics single-cell ATAC and RNA multiome sequencing data (also on 24hpf tails) and the Xenium experiment was highly successful in detecting tissue-specific expression patterns of the chosen genes with single-molecule resolution. DAPI and ribosomal RNA-based cell-body stains contributed to a seemingly quality cell-segmentation performed by the on-board Xenium software, allowing single-cell resolved clustering and annotation from the resulting cell-by-gene matrix. The resulting data presents several challenges for processing and analysis, with the immediate goal being visualization and quantification of differences along the entire ~800um embryonic trunk of six embryos (three replicates each in two different biological conditions) in a single experiment.
Speaker: Scott Lacadie -
09:31
Developing a high-content imaging workflow to investigate the impact of environmental chemicals on intestinal macrophages in zebrafish larvae 13m
The gut is one of the primary sites for exposure with chemicals and hence a vulnerable target for toxic effects. We hypothesize that intestinal exposure to such chemicals can trigger gut inflammation. Macrophages, key innate immune cells, represent a promising readout for detecting immunomodulatory effects following chemical exposure. To investigate this hypothesis, we developed an open-source workflow for 3D high-content image processing and analysis of transgenic zebrafish larvae expressing mCherry under the macrophage-specific promoter mpeg1. We used a 96-well setup to acquire 3D confocal images of zebrafish larvae, which were processed through a semi-automatic workflow. The intestine is manually defined in transmitted light images using an ImageJ macro. This anatomical region was then mapped onto corresponding fluorescence channels for precise quantification in python. To further streamline the process, we are training a 3D nnU-Net for semantic segmentation of the larval body and gut based on partially annotated volumes. Preliminary results from the beta-version of our workflow demonstrate that we can detect changes in macrophage numbers following exposure to reference chemicals, such as dextran sodium sulfate and trinitrobenzene sulfonic acid. Our approach addresses natural biological variability by correcting for the inherent differences in macrophage distribution and fluorescence between individual larvae. Our workflow involves the detection of cells across both the whole body and the intestine of each larva, enabling the calculation of a ratio between intestinal and whole-body cell counts. This normalization strategy helps correct for individual differences in overall fluorescence and ensures that detected changes in gut macrophage numbers truly reflect inflammatory responses rather than natural variability. However, the development of this workflow is time-consuming and non-linear, so the involvement of various colleagues and experts is crucial for success. In the workshop, we would like to present the workflow in its semi-automated beta version and discuss possible streamlining measures to improve the workflow.
Speaker: Elena Katharina Nicolay (Helmholtz-Zentrum für Umweltforschung GmbH UFZ) -
09:44
Leonardo: a toolset to remove sample-induced aberrations in light sheet microscopy images 13m
Selective plane illumination microscopy (SPIM, also called light-sheet fluorescence mi-croscopy) is the method of choice for studying organ morphogenesis and function as it permits gentle and rapid volumetric imaging of biological specimens over days. In inho-mogeneous samples, however, sample-induced aberrations, including absorption, scat-tering, and refraction, degrade the image, particularly as the focal plane gets deeper into the sample. Here, we present Leonardo, the first toolbox that is able to resolve all sam-ple-induced aberrations by using two major modules: (1) DeStripe removes the stripe ar-tifacts in SPIM caused by light absorption; (2) FUSE reconstructs one single high-quality image from dual-sided illumination and/or dual-sided detection while eliminating optical distortions (ghosts) caused by light refraction. The efficacy of Leonardo is validated on a wide range of biological systems, from minimally invasive experiments on sensitive spec-imens, for example, zebrafish embryos and optically opaque larval zebrafish, to immuno-labeling of wild-type mouse bodies up to roughly 2 centimeters. We publish a napari-based graphical user interface (GUI) and model code so the SPIM community can apply and improve Leonardo to advance imaging of inhomogeneous and complex specimens.
Speaker: Yu Liu (Technical University of Munich) -
09:57
Low-cost multiperspective imaging of zebrafish 13m
Automated multi-perspective imaging of zebrafish larvae allows researchers to accelerate analysis without the arduous task of repositioning specimens under a microscope. Systems, such as the VAST BioImager facilitate this, but they are expensive and lack adaptability for specific laboratory needs. Additionally, the glass capillary size used (600-700 µm) restricts the size of the larvae, which can be imaged. Here, we introduce a low-cost, open-source solution for automated multi-perspective imaging of zebrafish larvae.
Our 360° scanner captures images of zebrafish in a quartz glass capillary with an inner diameter of 3 mm while moving the camera around the stationary capillary. The capillary is housed in an imaging chamber filled with glycerin and sealed with a glass plate perpendicular to the camera. This setup minimizes image distortion. Moreover, because the refractive index of glycerin (n=1.475) and quartz glass (n=1.46) are nearly identical, the glass capillary becomes virtually invisible, reducing imaging interference. The imaging chamber is connected to the system via two pneumatic connectors, allowing easy interchangeability, for instance, to accommodate capillaries of different sizes to image zebrafish larvae at various developmental stages.
We use a Raspberry Pi HQ camera with a resolution of 4056 x 3040 pixels, equipped with a telecentric lens, which can be exchanged for lenses of various magnifications. The camera is mounted on a motorized rail, allowing it to move to the desired focal plane and enabling extended-depth-of-field imaging for sharp, high-depth images.
Our system predominantly uses 3D-printed components and readily available standard parts to maintain affordability. A graphical user interface enhances usability, offering options to image the zebrafish larvae at preset angle intervals for a full rotation or from a specific angle. Our next step is to automate the positioning of the specimens within the imaging chamber.
Speaker: Nathalie Klug (Institute for Automation and Applied Informatics - Biomedical Engineering & Robotics)
-
09:05
-
10:10
→
10:25
Coffee Break 15m
-
10:25
→
11:30
Talks 6-10
-
10:25
Zebrafish imaging workflows with Flamingo and Leonardo 13mSpeaker: Jan Huisken
-
10:38
Label-efficiency using self-supervised learning for classification of zebrafish embryo phenotypes in toxicity evaluations 13m
Machine Learning-based phenotype classification of zebrafish embryos enables fast and reproducible assessments for toxicity evaluation. However, acquiring large amounts of labeled data can be expensive, time-consuming and requires biological expertise. Based on the publicly available EmbryoNet dataset, we explore the impact that less available labeled data has on the performance of machine learning models and how leveraging unlabeled data using self-supervised pre-training can help in reducing the loss of performance. Furthermore, we investigate different data augmentation strategies to improve the visual representations obtained from self-supervised methods. We expect that the presented method will improve results for a wide range of tasks involving zebrafish embryos, especially when labeled data is scarce.
Speaker: Thomas Lautenschlager (KIT - IAI - ML4TIME) -
10:51
High Content Screening in zebrafish embryos using automated image and video analysis – advancements and challenges 13m
Zebrafish embryos are frequently used as an experimental model in the assessment of toxic effects. They are considered as alternatives to animal testing, since early life stages are not considered as “protected” stages by recent European regulations on animal welfare. An important read-out in effect assessment of chemicals in the zebrafish embryo is the assessment of morphological alterations. Typically, this has been conducted by microscopical observation and user-based scoring. This resulted in a bias in the effect assessment and difficulties to derive effect concentrations for chemical effect in a non-cumulative way for individual phenotype. To derive an unbiased quantitative assessment of morphological phenotypes we developed the software FishInspector (freely available via https://codebase.helmholtz.cloud/ufz/tb3-cite/biotox/FishInspector/-/tree/v1.87). By initial user annotation of the coordinates of structural features such as contour, eye, yolk, otoliths, mouth tip, notochord, pericard, fins and swimbladder in lateral and dorsoventral position, appropriate models were trained for an automated detection. The coordinates of the detected features were then used to derive quantitative metrics for concentrations response analysis, such as eye size, body lengths or otolith-eye distance (and many more). The first versions of FishInspector used models trained in MATLAB, based on a SegNet architecture with a pretrained VGG-19 encoder from the Deep Learning Toolbox. However, these models required downsizing of images and the derived annotations required relative intense user editing. Therefore, we revised the FishInspector software to incorporate PyTorch-based models trained in Python. The High-Resolution Network (HRNet), a state-of-the-art segmentation model, was integrated to address the multilabel segmentation task. While FishInspector is still coded in Matlab it is now interacting with a Python environment to annotate the structural features. An accessory software (MultiLabelFishTrainer) has been developed for a user-friendly training of Python models. Furthermore, we established a software (FishCardiologist) to analyse short 15-second videos from lateral position in order to extract the heart rate and to obtain an image of the cardiovascular system based on video frame subtractions. By combining these image analysis tools with the assessment of embryonic movement patterns, over 200 chemicals have been analysed so far and examples of a comparative analysis will be demonstrated. In the zebrafish workshop we would like to discuss major challenges in image analysis, such as assessment of quantitative analysis of complex structures (e.g. cardiovascular system) and the training of additional models.
Speaker: Stefan Scholz (UFZ - ETOX) -
11:04
Challenges in the automated analysis of zebrafish startle response behaviour 13mSpeakers: Gaelle Hayot (KIT), Thomas Dickmeis (KIT)
-
11:17
A Research Data Management (RDM) framework for High-Content Screening (HCS) bioimaging and video data of zebrafish (Danio rerio) embryos 13m
In recent decades, high content screening (HCS) assays using zebrafish (Danio rerio) embryos have become increasingly common in international research laboratories. Several of these assays involve the acquisition of large datasets in a few hours or the acquisition of video data using automated recording devices. After analysis, this data can aid in the understanding of cellular processes and support drug development testing. It is clear that such approaches produce a significant amount of information, ranging from raw data in various file formats (e.g., AVI, TIFF, JPG) to analysis results that require proper metadata annotation and storage. These files need to be linked to the fish embryo exposure assay (FET) results for proper context understanding. In addition to the importance of data storage for these files, which can be up to several terabytes in size, it is also crucial to consider the reusability of analysis pipelines according to FAIR (Findable, Accessible, Interoperable, and Reusable) standards. It is evident that research data management (RDM) for this specific data type presents a significant RDM challenge.
In this work, we showcase our in-house strategy for managing and storing zebrafish larvae images and videos. This framework comprises various data infrastructures and databases that can be interconnected using Workflow Management Systems (WMS). Unique sample ID links from the Integrated Effect Database for Toxicological Observations (INTOB) - a comprehensive data management tool for zebrafish effect data - can be linked to OMERO, which is used to store HCS bioimaging data and related metadata. Furthermore, we use the Helmholtz data infrastructure, including dCache/Infinite Space, to store video data and more space-intensive image formats. Finally, we developed KNIME and Galaxy pipelines to retrieve and analyze the data in a reproducible manner. This approach enables us to tackle the complexity of metadata, ensure HCS data integrity, and facilitate internal collaboration.Speaker: Riccardo Massei
-
10:25
-
11:30
→
11:55
Open Discussion
-
11:55
→
12:00
Close OutConveners: Christian Pylatiuk, Stefan Scholz (UFZ - ETOX)
-
09:00
→
09:05
