You can't always get what you want? - Yes, you can!
What is currently a crucial bottleneck in your community? What can we do to help you make spectacular progress? Which task, if solved, would make your scientific work easier on a daily basis? Which task, if solved, would have a enormous impact for you?
We are interested in your everyday problems as well as in questions that you have been putting aside for years. Making others aware of your research problems and bringing together experts from different fields could be the missing spark to solve your problems.
This is a unique opportunity to engage in discussions with other people interested in imaging without the need to prepare a poster or presentation. Or it can complement your poster presentation or talk by fostering a lively and fruitful discussion.
Session format: At the beginning you give a super-short pitch on your topic (1min). This pitch round is followed by a 45-minute open discussion in self-selected groups. Let's see if someone has THE idea for a solution to YOUR task.
Don't hesitate: no image is too big, no task too small!
Send your question and a short description so that we can plan to: contact@helmholtz-imaginge.de, subject: BYOIC and let the community work on your problems.
Examples
Quantitative Analysis of Mitochondrial Changes in Patients with Alcohol-Related Liver Disease (ARLD)
Background: Research indicates that mitochondrial dysfunction may play a crucial role in the progression of liver diseases associated with excessive alcohol use. Alterations in the number and size of mitochondria have been observed in liver cells as the disease advances.
Data Available: The dataset consists of high-resolution electron microscopy images of liver cells from patients at various stages of ARLD. These images offer detailed views of mitochondrial structures.
Objective: Our objective is to develop a quantitative method to measure mitochondrial changes, both in number and morphology, and to correlate these changes with the progression of liver disease related to alcohol use.
Technical Challenge: The challenge is to design an image processing algorithm that can accurately identify and measure mitochondria in electron micrographs. The difficulty includes handling the variability in mitochondrial appearance and differentiating them from other similar-looking cellular structures.
Expected Outcome: The development of a precise measurement technique will enhance our understanding of the mechanisms underlying ARLD and could lead to better-targeted treatments.
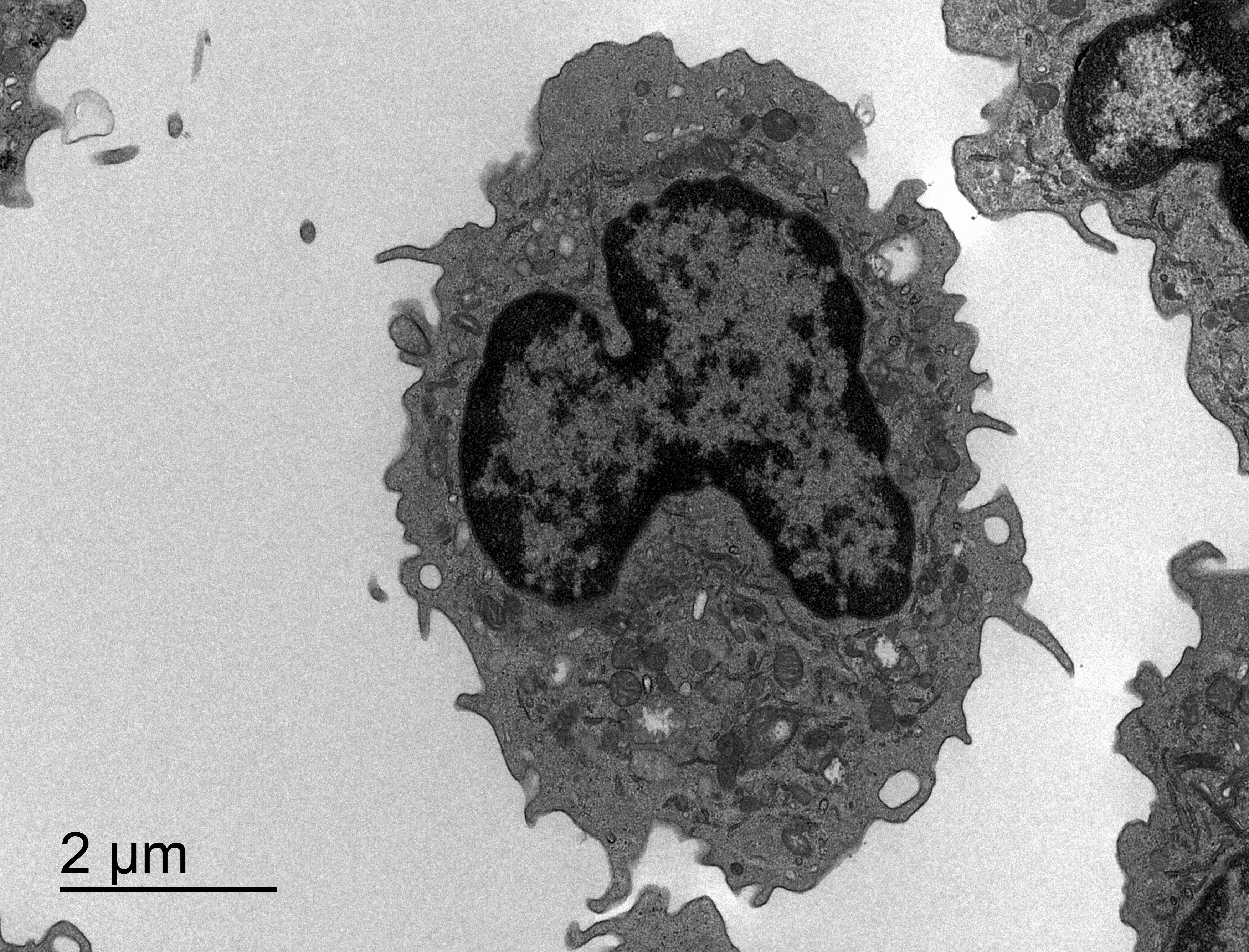 | 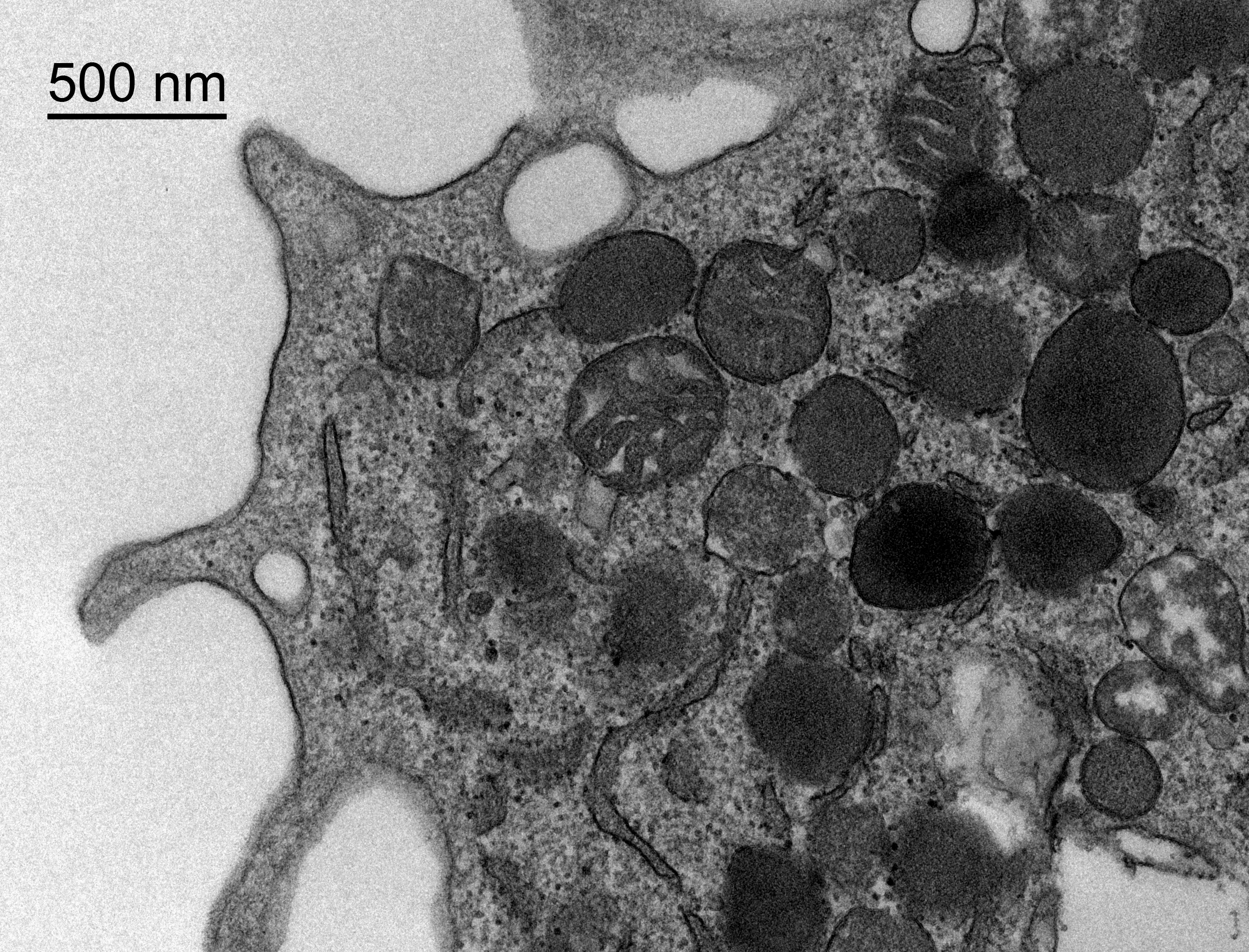 |
Images: Dr. med. Ingrid Wei Zhang, Department of Hepatology and Gastroenterology, Charité Berlin
Segmentation and Classification of Cell-Cell Contacts in Fluorescent Microscopy Images
Background: Cell-cell junctions play pivotal roles in tissue architecture, exhibiting variations across tissues and undergoing dynamic changes. Endothelial junctions are particularly vital for maintaining stability while facilitating cellular rearrangements. A deeper comprehension of cell-cell junctions holds promise for uncovering novel biological insights.
Data Available: The dataset comprises high-resolution fluorescent microscopy images depicting endothelial cells, with emphasis on cell-cell junctions (highlighted via VE-Cadherin staining). Furthermore, approximately 10k patches showcasing various junction types have been meticulously annotated, categorized into five groups: "straight," "thick," "thick/reticular," "reticular," and "fingers."
Objective: Our objectives are twofold. Firstly, to refine the segmentation of cell-cell junctions utilizing cellpose cell segmentation masks, thereby accurately delineating the boundary of the cell-cell junction region. Secondly, to classify the cell-cell junctions into one of the aforementioned five profiles by leveraging the annotated patches. Additionally, we aim to extend our analysis to include epithelial cells and cardiomyocytes. We provide a small annotation tool, functioning as a Discovery Tool that could aid in this endeavor by facilitating the identification and annotation of cell-cell contacts in these cell types.
Technical Challenge: The technical challenge herein is twofold. Firstly, it involves the design or enhancement of a semantic segmentation algorithm tailored for cell-cell interactions. Secondly, it necessitates the development of a classification algorithm grounded on handcrafted features derived from cell morphology and polarity to discern junction profiles. This entails utilizing the hand-annotated patches to facilitate predictions per junction area. Features extraction can be enabled through Polarity-JaM.
Expected Outcome: This endeavor will facilitate an automated approach to classifying cell-cell interactions, thereby advancing our comprehension of biological processes governing cell remodeling throughout development, homeostasis, and under pathological conditions.

