Speaker
Description
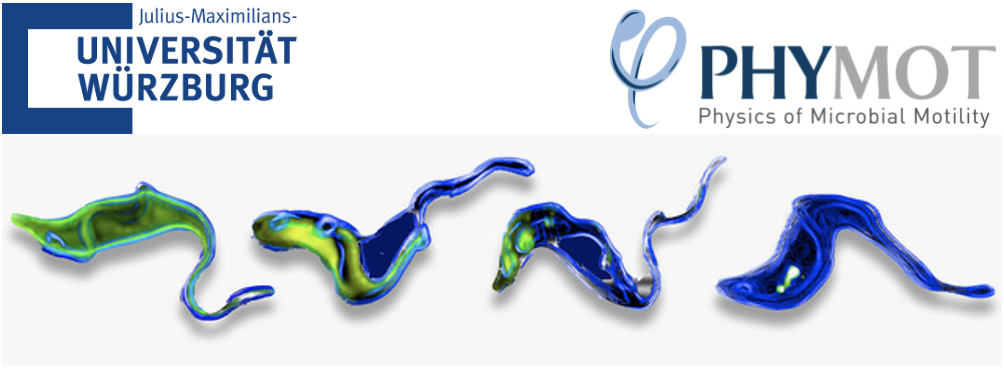
We investigate motility of the protozoan Trypanosoma, using numerical simulations. We have established a trypanosome model (see Fig. 1), which is motivated by experimental observations [1, 2] and builds upon the previously proposed model [3, 4]. The cell body is represented by a set of vertices which are distributed homogeneously on a pre-defined elongated surface and form a triangulated elastic network of springs. The network model also incorporates bending rigidity, and area and volume conservation constraints. For parasite propulsion, a flagellum is attached to the cell body. The flagellum is constructed from four parallely placed filaments, two of which are embedded into the body, and the other two are used for the generation of a propagating bending wave [5]. Flagellum beating leads to a deformation of the body and generates propulsion. We study the behavior of this model for different body stiffnesses, beating frequencies,wavelengths, and amplitudes. The simulations achieve values for the swimming velocity
and the body rotation around its swimming axis, which are at the same order of magnitude as experimental measurements. The trypanosome model is flexible enough and can be adapted to reproduce the behavior of different trypanosome species. This model will be used to investigatetrypanosome locomotion in blood stream.