- Indico style
- Indico style - inline minutes
- Indico style - numbered
- Indico style - numbered + minutes
- Indico Weeks View
Workshop on Digital Bioeconomy: Convergence towards a bio-based society
→
Europe/Berlin
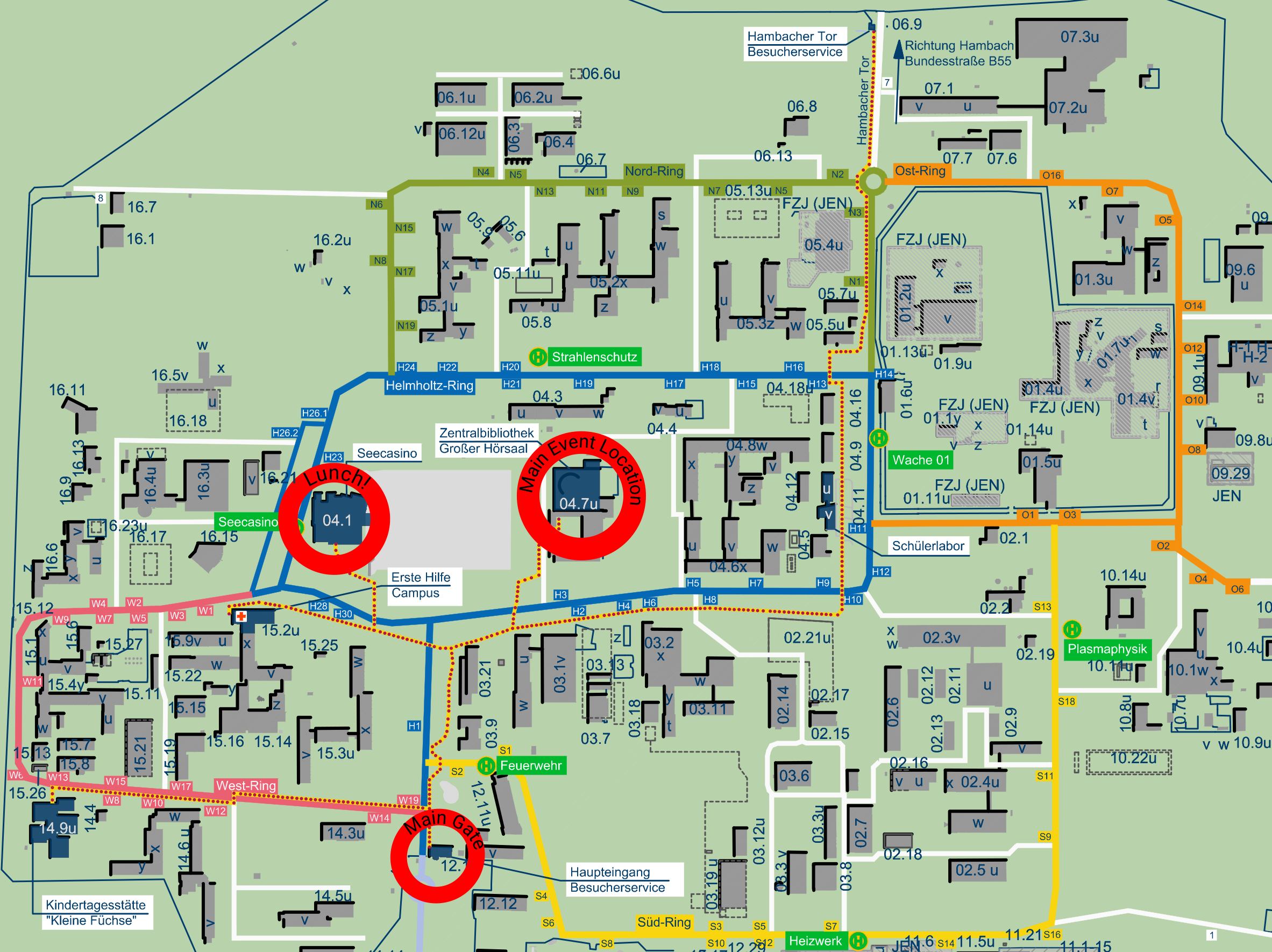
Lecture hall of the central library (Forschungszentrum Jülich)
Lecture hall of the central library
Forschungszentrum Jülich
Forschungszentrum Jülich GmbH
Wilhelm-Johnen-Straße
52428, Jülich
Germany
Description
Topics of the Workshop
• Data Standardization, Findability, and Reusability in Bioeconomy.
• Modeling and Data Science Approaches to Bioeconomy-Driven Biotechnology, Agrosphere, and Plant Research.
• Poster Session.
Scope of the Workshop
Every scientist and PhD student working in topics related to Bioeconomy.


-
-
09:00
→
09:15
Introduction / Welcome address 15mSpeakers: Peter Jansens (VS, Forschungszentrum Jülich GmbH), Holger Gohlke (IBG-4, Forschungszentrum Jülich GmbH)
-
09:15
→
09:45
FairAGRO in a nutshell - making agrosystem domain data fair and sustainable 30m
Here, we will present the NFDI FairAGRO dedicated to agrosystem data. These data require an integrated systems perspective to develop sustainable crop production systems, consider interactions between agriculture and the environment (e.g., plants, soil, microbiota), and relationships between scales (space, time, and organisms). Special features of agrosystem research data are geospatial information which might be coupled to data collected on private land (cropland, pasture). Accordingly, data protection aspects must be taken into account here.
The goal of the community-driven FairAGRO consortium is to provide researchers with a FAIR and quality-assured research data management (RDM) for generating, publishing, and accessing research data, to provide innovative and user-friendly RDM services, and to create modern data science methods for advancing agrosystems research. Using a Use case driven approach FairAGRO addresses key RDM challenges and contributes to the implementation of standards and services for selected research data infrastuctures. We will give an overview of FairAGRO the challenges in agrosystems research and FairAGRO's approach.Speaker: Björn Usadel (IBG-4, Forschungszentrum Jülich GmbH) -
09:45
→
10:15
EnzymeML – a data exchange format for biocatalysis and enzymology 30m
The design of biocatalytic reaction systems can be quite intricate, as numerous factors can impact the estimated kinetic parameters, including the enzyme itself, reaction conditions, and the chosen modelling method. This complexity can make reproducing enzymatic experiments and reusing enzymatic data challenging. EnzymeML 1 was created as an XML-based markup language to address this issue, enabling the storage and exchange of enzymatic data, including reaction conditions, substrate and product time course, kinetic parameters, and kinetic models. This approach makes enzymatic data accessible, findable, interoperable, and reusable (FAIR). Furthermore, the EnzymeML team and community have developed a toolbox that helps researchers report on experiments, perform modelling tasks, and upload EnzymeML documents to data repositories. The usefulness and feasibility of the EnzymeML toolbox have been demonstrated in various scenarios by collecting and analysing the data and metadata of different enzymatic reactions 2 . EnzymeML is a seamless communication channel that
connects experimental platforms, electronic lab notebooks, tools for modelling enzyme
kinetics, publication platforms, and enzymatic reaction databases. EnzymeML is an open
standard and available via https://github.com/EnzymeML and http://enzymeml.org.Speaker: Jan Range (University Stuttgart) -
10:15
→
10:45
Enhancing Bioeconomy Research Through FAIR Data Management: The Development and Application of LISTER for Efficient Metadata Extraction 30m
The importance of scientific methods, code, and data availability for replicating experiments is vital. This is particularly true in the context of the bioeconomy, where research data should be made available following the FAIR principle (findable, accessible, interoperable, and reusable). To facilitate this, we have developed LISTER, a solution designed to create and extract metadata from annotated, template-based experimental documentation, avoiding manual annotation effort for the experiment data generated during the research activities. LISTER is designed to integrate with existing platforms, using eLabFTW as the electronic lab notebook and the ISA (investigation, study, assay) model as the abstract data model framework. It consists of four components: an annotation language for metadata extraction, customized eLabFTW entries to structure scientific documentation, a "container" concept in eLabFTW for easy metadata extraction, and a Python-based app for semiautomated metadata extraction. The metadata produced by LISTER can be used as a base to create or extend domain-specific controlled vocabulary and ontologies, enhancing the value of published research data when applied. This is particularly relevant in the bioeconomy sector, where data-driven insights can drive sustainable solutions for global challenges such as food security and climate change. LISTER outputs metadata in machine-readable .json and human-readable .xlsx formats, and Material and Methods (MM) descriptions in .docx format that could be used in a thesis or manuscript. The extracted metadata and research data are hosted on DSpace, a data cataloging platform. We have successfully applied LISTER to computational biophysical chemistry, protein biochemistry, and molecular biology. We believe our concept can be extended to other areas of the life sciences, including those relevant to the bioeconomy, such as plant science research.
Speaker: Fathoni Musyaffa (University Düsseldorf) -
10:45
→
11:45
Poster session: Poster session/Coffee break
-
10:45
Amine transaminase engineering based on constraint network analysis and machine learning 1h
Amine transaminases (ATAs) are important enzymes for the production of chiral amines in the pharmaceutical and fine chemical industries[1]. However, the application of ATAs on novel substrates in an industrial setting usually still requires significant protein engineering efforts to optimize their structural and catalytic properties.
Intending to provide novel computational tools for in silico ATA engineering, we use our in-house software Constraint Network Analysis (CNA)[2] to study protein rigidity at the atomic level and map changes in protein rigidity during simulated heating to experimental measurements of thermostability. Additionally, we can identify structural weak spot residues, which have been shown to provide an increased likelihood for stability improvements upon mutation in other enzymes[3].
When comparing the predicted thermostability of 44 ATA variants to their experimentally measured temperature of half-inactivation (T50), we find two sub-populations of variants with significant, moderate correlations. These sub-populations arise from a single, highly stabilizing mutation, which is not identified by our out of the box application of CNA.We identified structural weak spot residues and applied the recently published machine learning model ProteinMPNN[4] with the goal to predict the most stabilizing mutations for every weak spot. These predicted novel variants are currently being tested experimentaly.
[1] F. Guo, P. Berglund, Green Chem., 2017, 19, 333-360.
[2] C. Pfleger, et al., J Chem Inf Model., 2013, 53, 1007-1015.
[3] C. Nutschel, et al., J Chem Inf Model. 2020, 60, 1568-1584.
[4] J. Dauparas, et al., Science. 2022, 378, 49-56.Speaker: Steffen Docter -
10:45
Design and Learn: Computational tools for guiding the development of a sustainable bioprocess 1h
Bio-based processes offer a solution to reduce reliance on polluting production processes and excessive land-use for food production. However, the design of such processes and effective microbial cell factories is hampered by the complexity of biological systems. The development of bioprocesses and microbial production strains can be accelerated by integrating concepts of resource allocation and condition-specific experimental datasets in metabolic models. To support the transition to a circular economy we focus on (1) the creation of innovative computational strategies for experiment design to streamline bioprocess development, and (2) the practical implementation of these methods on a hydrogen oxidizing bacteria to engineer a chassis capable of producing milk protein.
The first facet of our work involves the development of sophisticated computational methods to guide the design of experiments across all stages of bioprocess development, from initial concept to large-scale production. A central component is the establishment of an automated protein allocation model parametrization workflow. This workflow enables the seamless integration of diverse data sources, allowing for dynamic adjustments and optimization of protein allocation within the bioprocess.
The second aspect of our research focuses on the practical use of computational tools on Xanthobacter sp. SoF1, aiming to engineer this hydrogen oxidizing bacteria to produce milk protein from CO2 and N2. Our approach involves developing a metabolic model, making targeted genetic modifications, and optimizing pathways. By combining transcriptomics data with a protein allocation model, we design experiments to enhance milk protein production. This synergy between experimental design and computational models accelerates the optimization of bioprocess parameters for more efficient and sustainable production.
Speakers: Mr Aziz Ben Ammar (institute of Applied Microbiology, RWTH Aachen University), Samira van den Bogaard (institute of Applied Microbiology, RWTH Aachen) -
10:45
Engineering of an organic solvent tolerant esterase based on computational predictions 1h
Biocatalytic reactions in synthetic chemistry often require the presence of organic solvents to enhance substrate solubility or steer the reaction equilibrium towards synthesis. Given that enzymes have evolved in aqueous environments, the discovery of rare organic solvent-tolerant enzymes in nature is immensely valuable for elucidating the molecular mechanisms underlying their resistance. The esterase PT35 from Pseudomonas aestusnigri, which was found in crude oil contaminated sand samples, exhibit extraordinary activity and stability (Tm 49°C; t1/2 35 h) in the presence of 50% acetonitrile. Molecular dynamics (MD) simulations comparing PT35 with the organic solvent-sensitive structural homolog ED30 revealed a more pronounced hydration shell around PT35 due to its distinctive negative surface charge. We developed a mutagenesis strategy involving both enzymes, PT35 and ED30, that aimed to decrease or strengthen the hydration shell surrounding the enzyme by modifying its surface charge. Consequently, we generated PT35 variants with reduced negative surface charge and ED30 variants with enhanced negative surface charge. We successfully produced 10 variants for each enzyme, with 7 PT35 variants exhibiting decreased tolerance (e.g. Δt1/2 -37.2 h) and 8 ED30 variants displaying increased tolerance (e.g. Δt1/2 +25.6 h) compared to the wild type. Our findings underscore the potential of engineering an enzyme's surface charge as a means to boost its tolerance to organic solvents. Our next goal is to produce a ED30 variant with 7 mutations evenly distributed across the surface to reinforce the hydration shell around the entire enzyme.
Speaker: Lara Scharbert -
10:45
Engineering PET-degrading enzymes – targeting the energy barrier for PET binding 1h
In view of the worsening climate crisis and increasing plastic waste pollution, scientific interest in the development of an environmentally friendly enzymatic degradation mechanism for plastics is growing. However, the bottleneck in the industrial application of enzymes for plastic waste recycling is their insufficient activity and partial lack of stability under industrial conditions.
To this end, we investigated the binding behaviour of highly active PET-degrading enzymes to polyethylene terephthalate (PET). Adsorption to the PET surface could be captured by classical molecular dynamics (MD) simulations. However, the entry of PET into the active site associated with the formation of productive binding poses was presumably hindered by an energy barrier limiting the activity of the enzyme. Using Hamiltonian Replica Exchange MD (HREMD) simulations, we were able to overcome this barrier and unveil entry pathways leading to productive conformations using principle component analyses (PCA).
In addition to hindering intramolecular PET interactions, we identified amino acids that potentially hinder entry into the binding site based on free energy surface (FES) profiles of amino acid-PET interaction and evaluated the PET-degradation activity of respective variants experimentally.
These insights address current research gaps on the mechanism of PET-degrading enzymes through comprehensive computational analyses of the full binding pathway, which highlights the importance to address not only substrate binding, but also the entrance into the active site.
Speaker: Anna Jaeckering (Forschungszentrum Jülich) -
10:45
Exploring the Microbial Symphony - Unveiling the Dynamics of Biogas-Producing Communities 1h
Research on biogas-producing microbial communities focuses on uncovering the correlations and dependencies between process parameters and community compositions to optimize process stability and biogas output. The tremendous advances in High-Throughput (HT) sequencing technologies have shifted the study of microbiomes predominantly to metagenome research methods. Thousands of Metagenomically Assembled Genomes (MAGs) have been compiled from biogas microbiomes using metagenome assembly and binning approaches. Deep sequencing of biogas microbiomes has made it possible to reconstruct genomes from community members and predict how these communities adapt to specific conditions at the species level. In this context, MAG-based metabolic reconstruction has provided insights into the genetically determined metabolic potential of the previously less understood genus Limnochordia, which is predicted to be metabolically versatile and ubiquitously active in many pathways of the biogas process chain. Interestingly, Limnochordia appears resilient to varying process conditions, making it a compelling candidate for production applications as shown in this study. However, in most cases, physiological experiments to confirm metabolic predictions are not feasible, as isolates for many MAGs are unavailable. Therefore, culturomics is essential to determine the phenotypes of microorganisms represented by MAGs, complementing the functional profiling of microbiome members.
Speaker: Irena Maus (IBG-5) -
10:45
Open Source Tools and FAIR Data Formats for Mass Spectrometry Lipidomics 1h
The Lipidomics Informatics for Life Science (LIFS) consortium, part of the German Network for Bioinformatics Infrastructure (de.NBI), pioneers advancements in lipidomics workflows through the development of a comprehensive suite of tools.
LipidCreator facilitates targeted LC-MS/MS assay generation and seamlessly integrates with Skyline for method file generation and data processing on state-of-the-art mass spectrometers from major vendors. LipidXplorer enables vendor-agnostic processing of untargeted, shotgun lipidomics data, allowing users to define custom lipid fragment patterns and rules using the mass spectrometry fragment query language (MFQL). Goslin is a polyglot grammar-based parsing and conversion library, harmonizing and mapping different lipid nomenclatures to the latest endorsed by LIPID MAPS and the lipidomics standards initiative (LSI). Finally, LUX Score, extended by LIFS, and its successor, LipidSpace, offer qualitative lipidome comparisons and similarity calculations. LipidSpace further facilitates interactive quantitative comparison of lipidomes on matching structural hierarchy levels, serving as an effective tool for post-hoc analysis, comparison, and visualization of published lipidomics data.
Collaborating with HUPO-PSI MS, we introduced mzTab-M, a tabular reporting format now widely supported by metabolomics and lipidomics tools and repositories. Simultaneously, we are standardizing the mzQC data format to capture typical and user-defined QC metrics for mass spectrometry experiments in proteomics, metabolomics, and lipidomics.
In summary, the LIFS consortium's comprehensive suite of tools, including LipidCreator, LipidXplorer, Goslin, LUX Score, and LipidSpace, empowers high-throughput, quantitative lipidomics. Our commitment to FAIR data principles ensures seamless data handling, submission, and integration, furthering the accessibility and interoperability of lipidomics research.
Speaker: Nils Hoffmann (IBG-5, Forschungszentrum Jülich GmbH) -
10:45
Prediction of more thermostable ApPDC variants by Constraint Network Analysis and ProteinMPNN to enable an industrial application 1h
Carbon-carbon bond-forming reactions are essential for organic synthesis. In conventional petrochemical-driven approaches, these reactions rely on organic solvents and hazardous starting materials. In contrast, biocatalysts offer an alternative pathway based on second-generation feedstocks. In particular, pyruvate decarboxylase from Acetobacter pasteurianus (ApPDC) offers a sustainable alternative in the synthesis of chiral aromatic derivatives, facilitating the C-C bond formation between, e.g., benzaldehyde and pyruvate in aqueous solution. However, ApPDC exhibits insufficient thermostability for widespread industrial application.
Using molecular dynamics simulations, we generated a diverse conformational ensemble upon which we applied our in-house software, Constraint Network Analysis (CNA), to perform thermal unfolding simulations. These simulations unveiled weak spots within the protein structure. To validate our approach, we compare these identified weak spots with a previously described variant with a remarkable 12 °C enhancement in the melting point, which differs in 14 amino acid positions from the wild type. Our analysis successfully identifies 8 out of 14 specific weak spots within the ApPDC variant, underscoring the predictive capabilities of CNA in pinpointing thermal weak spots.
The predicted weak spots are substituted by mutations derived from ProteinMPNN that enhance the stability of ApPDC. Overall 5000 sequences with enhanced thermostability were generated. This research paves the way for accelerated progress in enzyme engineering for enhanced industrial applications.Speaker: Daniel Fritz Walter Becker (Heinrich-Heine-Universität) -
10:45
Quantum Mechanics/Molecular Mechanics (QM/MM) simulations: A modeling tool for Biomedicine and Biotechnology 1h
Molecular knowledge of enzyme-ligand complexes is crucial to understand their reactivity and thus design small molecules/protein mutations able to inhibit/modify enzyme activity. Complementing data science and other modeling approaches, Quantum Mechanics/Molecular Mechanics (QM/MM) simulations can provide further information on the enzymatic mechanism, thus facilitating such biomedical and biotechnological applications.
Here we exemplify this approach using as test case the unknown human gut bacterium mannoside phosphorylase (UhgbMP). Such enzyme is involved in the metabolization of eukaryotic N-glycans lining the intestinal epithelium, a factor associated with the onset and symptoms of inflammatory bowel disease. In addition, UhbgMP has been investigated for its potential to synthesize N-glycan core oligosaccharides very efficiently and less costly than by chemical means.
Using QM/MM simulations [1] we showed that the phosphorolysis reaction catalyzed by UhgbMP follows a novel substrate-assisted mechanism, in which the 3' hydroxyl group of the mannosyl unit of the substrate acts as a proton relay between the catalytic Asp104 and the glycosidic oxygen atom. Given the crucial role of the active site hydrogen bond network in the reaction and its conservation across mannoside-phosphorylases, the computed mechanism is expected to apply to other closely related enzyme families. Moreover, our simulations unraveled the conformational itinerary followed by the substrate along the reaction; such information can be used to design enzyme inhibitors. Future QM/MM simulations on the reverse phosphorolysis reaction also catalyzed by UhgbMP will provide hints to design protein mutations aiming at improving the biosynthetic yield of high added value products.
[1] https://doi.org/10.1021/acscatal.3c00451
Speaker: Dr Mercedes Alfonso-Prieto (Forschungszentrum Jülich) -
10:45
Seed-to-plant-tracking: Automated phenotyping and tracking of individual seeds and corresponding plants 1h
Seeds play a critical role in keeping continuity between successive generations of plants. However, it is still not fully understood whether, or to what extent, the variability of seed traits within plant species or genotypes in interaction with a changing environment has an impact on seed emergence (i.e. germination), early development and further performance of a plant. Here, we present the technology developed by IBG-2 to tackle these questions: our seed-to-plant-tracking approach includes 1) automated identification, measurement and sowing of individual seeds, 2) automated trait characterization of resulting seedlings and further plant growth, 3) harvest and storage of seeds for subsequent measurements in the next generation, 4) integration of data from distributed sources into a common database. The pipeline consists of the robotic system “phenoSeeder” (Jahnke et al. 2016), amended by an acoustic volumeter (Sydoruk et al. 2020), and the imaging platform “Growscreen” (Walter et al. 2007, Scharr et al. 2020). The main goal is to find correlations between seed traits and plant performance in all developmental stages. In combination with epigenetics, we aim to uncover how adverse conditions during plant development feed back on the variability of seed traits and early vigour of plants in the next generation. The identification of relevant plant traits and genotypes with promising properties under unfavourable environmental conditions will contribute to the sustainable intensification of plant production.
References:
Jahnke S, Roussel J, Hombach T, et al. (2016). Phenoseeder - a robot system for automated handling and phenotyping of individual seeds. Plant Physiology, 172:1358–1370.
Walter A, Scharr H, Gilmer F, et al. (2007). Dynamics of seedling growth acclimation towards altered light conditions can be quantified via GROWSCREEN: a setup and procedure designed for rapid optical phenotyping of different plant species. New Phytologist, 174:447–455.
Scharr H, Bruns B, Fischbach A, et al. (2020). Germination Detection of Seedlings in Soil: A System, Dataset and Challenge. In: Bartoli, A, Fusiello, A (eds) Computer Vision – ECCV 2020 Workshops. ECCV 2020. Lecture Notes in Computer Science, vol 12540. Springer, Cham.
Sydoruk V, Kochs J, van Dusschoten D, Huber G, Jahnke S (2020). Precise volumetric measurements of any shaped objects with a novel acoustic volumeter. Sensors 20:760.Speaker: Gregor Huber -
10:45
TopEnzyme significantly improves classification of enzyme function by incorporating computational enzyme structures. 1h
It is essential to understand target enzyme function for applications in biomedicine and biotechnology. A good method to predict the function of new enzymes is the classification through neural networks in combination with large structural datasets. To keep computational requirements feasible for these large systems, we created a more sophisticated representation of an enzyme than the sequence or fold while retaining local chemical information. This localized 3D enzyme descriptor improves enzyme function prediction compared to established methods in the field for GCNs.
For this project we developed TopEnzyme, a database of structural enzyme models created with TopModel and it is linked to the SWISS-MODEL and AlphaFold Protein Structure Database to provide an overview of structural coverage of the functional enzyme space for over 200,000 enzyme models. It allows the user to quickly obtain representative structural models for 60% of all known enzyme functions. We assessed the models we contributed with TopScore and found that the TopScore differs only by 0.04 on average in favor of AlphaFold2 models. We tested TopModel and AlphaFold2 for targets not seen in the respective training databases and found that both methods create qualitatively similar structures.
Testing the localized 3D descriptor on this database improves the F1-score up to 17% in enzyme classification tasks compared to fold representation methods. Furthermore, we implemented better GCNs, SchNett and DimeNetPP, for atom classification. This increases the performance by 13% and 16% on the enzyme classification task. Furthermore, we investigated the networks using GNNExplainer and found relational information more important when classifying residue-based objects, while chemical interactions are marked more important when classifying on atom objects. Our results demonstrate that a localized 3D descriptor is the better alternative to current reduced structure representations used in enzyme prediction networks.
Speaker: Karel van der Weg (Forschungszentrum Juelich) -
10:45
TransMEP: Transfer learning on large protein language models to predict mutation effects of proteins from a small known dataset 1h
Machine learning-guided optimization has become a driving force for recent improvements in protein engineering. In addition, new protein language models are learning the grammar of evolutionarily occurring sequences at large scales. This work combines both approaches to make predictions about mutational effects that support protein engineering. To this end, an easy-to-use software tool called TransMEP is developed using transfer learning by feature extraction with Gaussian process regression. A large collection of datasets is used to evaluate its quality, which scales with the size of the training set, and to show its improvements over previous fine-tuning approaches. Wet-lab studies are simulated to evaluate the use of mutation effect prediction models for protein engineering. This showed that TransMEP finds the best performing mutants with a limited study budget by considering the trade-off between exploration and exploitation.
Speaker: Birgit Strodel (Forschungszentrum Jülich)
-
10:45
-
11:45
→
12:15
Learning more with less: Hybrid Models, Transfer Learning, and Calibration Designs for Efficient Knowledge Transfer in Industrial Settings 30m
Industrial process development often follows a repetitive pattern, relying on the same experimental approach for each new product, despite incorporating knowledge into process templates (e.g. in form of platform processes which are widely spread in the (bio)pharmaceutical industry). While risk analysis under the Quality by Design (QbD) paradigm enables some vertical knowledge transfer, capturing the functional process behavior remains a challenge. Current efforts to develop generic mechanistic models face limitations due to their rigid structure, prompting the integration of machine-learning approaches for improved variability handling. This contribution explores the application of transfer learning in hybrid models, utilizing dummy variables, embeddings, and meta learners to transfer knowledge between molecules. Additionally, we present the concept of calibration designs, demonstrating how experiments can efficiently uncover process behavior for new molecules. Finally, we emphasize the necessity of establishing a standardized, self-learning environment for all stakeholders to effectively leverage model-derived knowledge in process development and tech transfer.
Speaker: Moritz von Stosch (DataHow AG) -
12:15
→
12:45
Facilitating Research Data Management for Fundamental Plant Science 30m
The evolution of digital methods and the broad availability of high throughput methods makes efficient research data management an essential necessity. This applies in particular to the field of fundamental plant science. Within the NFDI consortium DataPlant we rely on the ISA-Tab standard to annotate research data. Well annotated research data objects are called Annotated Research Context (ARC) and contain the actual research data as well as curated metadata describing its content. The specific benefit of using ARCs lies within their versioning and tracking of provenance, relying on Gitlab instances as federated repositories. DataPLANT offers various web-based services and tools to facilitate the handling of research data for scientists.
Speaker: Jens Krüger (Eberhard Karls Universität Tübingen) -
12:45
→
13:00
Group Photo 15m
-
13:00
→
14:15
Lunch 1h 15m
-
14:15
→
14:45
Modeling soil functions for a sustainable bioeconomy 30m
Soils occupy a central position in the ecosystem and provide essential functions for both, humans and the environment. On the one hand, they are used to produce biomass for human nutrition and animal feed, but also for bioenergy, and the production of various industrial materials. In addition, they are the largest terrestrial carbon reservoir, filter and store water, recycle nutrients and provide a habitat for an immense diversity of organisms. For many years, agriculture was almost exclusively production-oriented. Today, climate change, biodiversity loss and a number of other environmental problems are calling for a change to more sustainable production taking into account the entire bundle of soil functions.
The BODIUM model (König et al., 2023) is a systemic soil model which aims to simulate the effect of changing agricultural management practices on soil functions such as yield, water storage and filtration, nutrient recycling, carbon storage and habitat for biodiversity. For this purpose, the influence of crop rotation, soil cultivation, fertilization as well as the effect of a changing climate is taken into account site-specifically. A version of this model, the BODIUM4Farmers, is intended to serve farmers as an on-site decision support tool for long-term planning of soil management measures in response to actual economic and ecological requirements. In this presentation, we introduce soil as a central resource for a sustainable bioeconomy and provide an overview about the BODIUM and BODIUM4Farmers models.Speaker: Ute Wollschläger (Helmholtz Centre for Environmental Research) -
14:45
→
15:15
Getting value out of digitalization in biotech R&D 30m
The ability to use data and predictive computational technologies holds a large promise for developing the next generation of solutions in industrial and pharma-based biotech. Large language model technology applied to protein sequences combined with application-directed AI oracles have made spectacular progress in some cases, and there is a growing awareness that data-based design could indeed revolutionize the discovery process for many protein-based solutions.
In the present talk I will outline the view of these technologies from an industrial biotech view, and discuss the dilemmas that we have encountered over the years in Novozymes and now Novonesis in harnessing the power of computational biology and machine learning. I will discuss technical, practical and organizational aspects of how one can formulate the setup of a data-driven R&D organization and discuss successes and failures of our endeavors.
Finally, I will speculate on the future, whether it will resemble the past, and muse on how the computational biology community could chose to invest its resources for maximum impact in the 2020s and beyond.
Speaker: Jens Erik Nielsen (Novozymes A/S) -
15:15
→
15:45
AI-powered protein engineering: the next frontier in biocatalyst development 30m
Enzymes are a transformative tool for the path towards a digital bioeconomy due to their ability to catalyze diverse biochemical processes in sustainable and adaptable ways. Protein engineering plays a pivotal role in advancing the development of such biocatalysts with applications spanning biotechnology, biomedicine, and life sciences. The advent of protein-directed evolution, recognized with the 2018 Nobel Prize, has enabled the customization of enzyme functions for a broad range of new-to-nature applications. Despite significant progress, challenges persist due to a limited understanding of protein function and the complex multi-factorial optimization problem inherent in enzymes and proteins. Recent advancements, however, integrate machine learning (ML) techniques to address these challenges. Exazyme focuses on leveraging ML algorithms to enhance protein functionalities, including enzyme properties.
Our methodology employs a two-step ML model application. Initially, our models proficiently predict protein sequence-to-function mappings based on functionally assayed sequence variants, requiring minimal reliance on detailed mechanistic or structural data. This approach has proven particularly useful in low data regimes, where extensive screening is not possible. Subsequently, we utilize these predictions in a Bayesian optimization framework to guide the selection of candidates for experimental validation, enabling simultaneous optimization of multiple parameters, such as stability, catalytic speed, and substrate specificity. A noteworthy accomplishment of our research lies in the superior performance of our prediction algorithms, consistently outperforming current state-of-the-art methods, across various datasets and benchmarks. Practical validation of our algorithms was demonstrated through successful protein engineering campaigns, enhancing the functionality of complex enzymes from diverse families including carboxylases, hydrogenases, and phosphohydrolases.
Our results underscore the capacity of ML methods to expedite the processes of directed evolution and rational design of proteins. By efficiently predicting and selectively identifying sequences with improved properties, these methods leverage existing sequence variant data to advance the field of protein engineering, and thereby a digital bioeconomy.
Speaker: Jelena Ivanovska (Exazyme GmbH) -
15:45
→
16:15
Coffee break 30m
-
16:15
→
16:45
Computer Aided Enzyme Discovery and Engineering for Industrial Biocatalysis: From physics based MD to Machine Learning and AI 30m
Enzyme engineering by directed evolution allows the optimisation of virtually any enzyme into a powerful industrial catalyst, which can be conveniently applied under process conditions. However, due to time and cost limitations, enzyme discovery from (meta)-genomic diversity often fails to deliver high performance catalysts ready for industrial applications and enzyme variant screening under process conditions can only cover a very limited protein sequence space.
In order to overcome this limitation in Enzyme Discovery & Engineering, Enzymaster has developed the BioEngine®, a proprietary directed evolution platform with integrated computational enzyme identification/enzyme engineering toolbox BioNavigator®. This enables us to cover much larger sequence space using machine learning/AI for combining activity, 3D-structure and sequence information in our EM 2 L-platform for predictive in-sillico screening.
In this presentation I will show, how Enzymaster is utilizing it`s digital toolbox in order to speed up our smart enzyme evolution platform BioEngine® by computer aided enzyme engineering. We combine high throughput screening and next generation sequencing for fast and reliable data generation with AI and machine learning approaches for virtual enzyme identification and screening.
Enzymasters aim is to deliver practical solutions enabled by a powerful combination of experimental and virtual screening to overcome the hurdle from idea to product in a future data driven bioeconomy.Speaker: Marco Bocola (Enzymaster Deutschland GmbH) -
16:45
→
17:15
Soil sensing for soil health assessment – a contribution to digital bio-economy. 30m
Soil is essential for crop production and has other important functions with regard to ecosystems and human health. Proper soil management requires detailed knowledge of soil state variables and processes. However, soil investigation is challenging because soil is a very complex and heterogeneous medium with high spatio-temporal variability. Traditional laboratory based soil analysis is expensive and time-consuming. Soil sensing, in particular proximal soil sensing, promises to provide soil data with high resolution at minimum costs. The talk will present the state-of-the art in proximal soil sensing and discusses its prospects and limitations.
Speaker: Robin Gebbers (University Halle-Wittenberg) -
17:15
→
17:30
Closing remarks 15mSpeaker: Holger Gohlke (IBG-4, Forschungszentrum Jülich GmbH)
-
09:00
→
09:15
